Machine Learning for Life Sciences¶
Follow along at: 5x5x5x5.github.io/Machine_Learning_Life_Sciences
Definitions sure to annoy serious practitioners¶
Machine Learning - Using algorithms and computation to generalize from data
Life Sciences - Anything that wiggles from 20nm to 30m in length
Most of the subjects I will touch on are incredibly deep and worthy of their own talk. Thankfully, the Research Triangle Analysts have already given some of them.
- Tim Hopper on scikit-learn
- Python Data Science with Pandas
- Random Forests and Boosted Trees, subtitled The Entropy Of My Entropy Is My Friend by Dan Kelly
Opportunities in the Life Sciences¶
- Research frontier is relatively close
- New developments aimed at reducing costs and increasing reproducibility
- Science Exchange
- Riffyn
- Emerald Cloud Lab
- Transcriptics
- More data to feed the Data Scientists
- New approaches open up new avenues for data analysis
Three classification problems with three techs¶
- classification (QSAR) using scikit-learn in Python
- next gen sequencing RNA-seq MLSeq classification example in R
- diagnostic image analysis with ConvNets in Python using Theano
Remote Laboratory Services facilitated by Science Exchange 
Protocol Standardization and Optimization by Riffyn¶
Quality systems engineering for research methods
Design of experiments analysis
Interview with the Founder of Riffyn

Automated Research Pipelines by Emerald Therapeutics and Transcriptic¶

What to do with all that data?¶
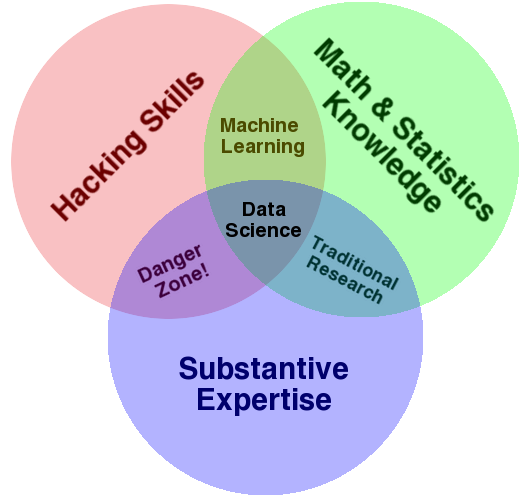
Highway to the Danger Zone?¶
- no statistical grounding
- no basis for claims
Fortunately, it requires near willful ignorance to acquire hacking skills and substantive expertise without also learning some math and statistics along the way. As such, the danger zone is sparsely populated, however, it does not take many to produce a lot of damage. - Drew Conway
Three classification problems with three techs¶
- classification (QSAR) using scikit-learn in Python
- next gen sequencing (standard R processing pipeline) RNA-seq
- diagnostic image analysis with ConvNets in Python using Theano
The emphasis here is on finding a common understanding of the vocabulary between life scientists and analysts; things like pipelines, dataframes and representations.
Classification in Machine Learning¶
A classic supervised learning problem.
Here is an excellent visualization of the process
1 Choose a representation
2 Train a classifier
- Holdouts
- Cross-validation
- Classifier selection
- Parameter search
3 Make predictions
4 Evaluation metrics
Quantitaive Structure Activity Relationship for Predictive Toxicology¶
Check out some chemicals¶
from rdkit import Chem
from rdkit.Chem import Draw
%matplotlib inline
m3 = Chem.MolFromSmiles('O=C1OC2=C(C=C1)C1=C(C=CCO1)C=C2')
fig3 = Draw.MolToMPL(m3)
smiles = ("O=C(NCc1cc(OC)c(O)cc1)CCCC/C=C/C(C)C", "CC(C)CCCCCC(=O)NCC1=CC(=C(C=C1)O)OC", "c1(C(=O)O)cc(OC)c(O)cc1")
mols = [Chem.MolFromSmiles(x) for x in smiles]
Draw.MolsToGridImage(mols)
Next Gen Sequencing - RNA-Seq¶
NGS Woodchipper
Dealing with 30X genome sized datasets initially
- lots of interesting statistics work around:
- multiple hypothesis testing
- assembly and annotation of nucleic acid sequences
- error correction
- not machine learning
Comparing RNA expression levels takes this from a big data problem back to another simple classification problem
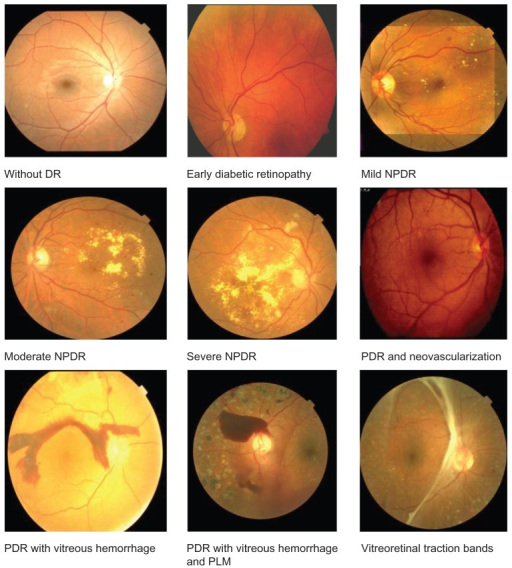
Diagnostic Image Analysis¶
Diabetic Retinopathy Competition recently from Kaggle.
The top 10 all used Convolutional Neural Nets
The winners in their own words¶
First place winner Ben Graham
Fourth place winners Julian De Wit & Daniel Hammack
Jeffrey De Fauw most lucid explination yet.
Ramon Peres got a PLOS publication out of his entry.
Introduce deep learning
Please do not get all Strong AI on me
Lends itself to parallel computation. GPUs are usefull for this.
Picture of simple net
Picture of architecture
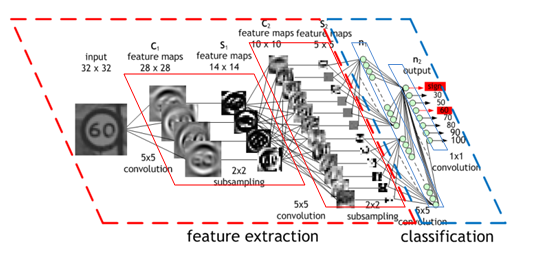
Example of python code with Theano
New opportunities come from tying together multiple models.
Tools for speaking the same language¶
Facility at the command line opens up a world of open source software
Version control -- git
Use AWS free tier¶
Groundhog Day for computing
yes, it's slow, but it's free
scripts coming to github
Thanks for listening. Next steps.¶
For the hackers
- Stay away from anything making claims that would involve the FDA or HIPAA or humans in general.
- There is enough going on in your yard or compost pile or maybe even your fridge.
- Check out TriDIYbio
For the employed
For the enthusiast
- Help me start the Research Triangle BioCoders
- Try to find public biological datasets and publish better tutorials and example analyses.
!jupyter nbconvert --to slides MLforLS.ipynb --post serve
[NbConvertApp] Converting notebook MLforLS.ipynb to slides [NbConvertApp] Writing 202636 bytes to MLforLS.slides.html [NbConvertApp] Redirecting reveal.js requests to https://cdn.jsdelivr.net/reveal.js/2.6.2 Serving your slides at http://127.0.0.1:8000/MLforLS.slides.html Use Control-C to stop this server Created new window in existing browser session. WARNING:tornado.access:404 GET /custom.css (127.0.0.1) 0.79ms WARNING:tornado.access:404 GET /favicon.ico (127.0.0.1) 0.47ms